Yet another oprofile tutorial
Recently it came as a surprise to me that many people don't know how to use oprofile efficiently when working on performance optimizations. I'm not going to duplicate the oprofile manual here in details, but at least will try to explain some basic usage.
A bit of theory
Oprofile does its magic by using statistical sampling. The processor gets interrupted at regular intervals (the interrupts happen after a certain amount of time has elapsed, or some hardware performance counter accumulated a certain amount of events) and oprofile driver identifies which code had control at that moment. The part of code which was 'lucky' to be interrupted by oprofile, gets an oprofile sample attributed to it. The parts of code which take a lot of execution time are naturally more likely to accumulate many oprofile samples. In fact, the amount of collected oprofile samples for some function tends to be directly proportional to the execution time taken by this function. This all is somewhat similar to Monte Carlo method.
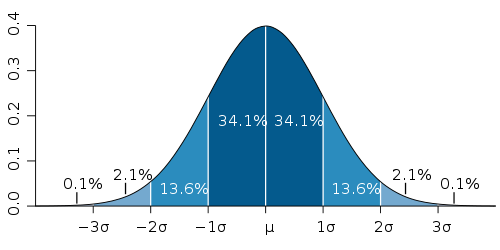
The collection of samples done by oprofile for each individual function is a Poisson process. Standard deviation for Poisson distribution is the square root of the number of samples. So the more samples got collected, the lower is the relative error. The following diagram shows the confidence intervals for normal distribution (because Poisson distribution is approximately normal for the large number of samples):
Using the 3-sigma rule, we can be fairly confident that the actual time spent in each function (measured in oprofile samples) is within 3*sqrt(N) interval for each function. Where N is the number of samples reported by oprofile for that function.
A simple profiling and code optimization workflow
Let's suppose that we have some small command line tool, which can be run to do something useful. And we want to optimize this tool to spend less time to do the same work. First of all, it makes sense to identify the parts of the program, which are the performance bottlenecks and can be optimized. This can be easily done using oprofile:
# opcontrol --deinit
# opcontrol --separate=kernel
# opcontrol --init
# opcontrol --reset
# opcontrol --start
# ./test-program
# opcontrol --stop
# opreport -l ./test-programGoing through all of the above steps will configure and start oprofile, then execute the program to be profiled (./test-program), then finally stop oprofile and show the profiling report (and this report contains exactly the information we want, and its interpretation is explained a bit in the next section). The opcontrol tool needs to be run as root or via sudo. It is also quite important to use --separate=kernel option. This option is described in details here, but basically it ensures that all the CPU activity happening in the kernel and in the shared libraries is also attributed to the test program and shown in the log.
After having oprofile report, it is only a matter of checking what parts of code are reported to take a lot of time, improving them and finally running oprofile again to verify the results. This process can be repeated multiple times. That's quite simple. Though there are two main cases when it may be difficult to interpret oprofile logs:
- Oprofile reports that just one large function (possibly even 'main') is taking most of the time.
- Oprofile reports a million of tiny functions, each taking only a small fraction of time.
In the former case it is a good idea to split the large function into a few smaller ones. If the large function is already calling some other functions which aree inlined, then naturally disabiling inlining will provide a bit more interesting profiling report. Another alternative is to use source annotation. But be sure to read about all the caveats in the oprofile manual. In the latter case, generating a callgraph may provide some insights. Some nice callgraph pictures can be generated by Gprof2Dot from the data collected by oprofile.
A real practical example
I'm going to use one of my old performance patches as an example. Oprofile report for the 'sbcenc' program looked like this before optimization:
samples % image name symbol name
26083 25.0856 sbcenc sbc_pack_frame
21548 20.7240 sbcenc sbc_calc_scalefactors_j
19910 19.1486 sbcenc sbc_analyze_4b_8s_neon
14377 13.8272 sbcenc sbc_calculate_bits
9990 9.6080 sbcenc sbc_enc_process_input_8s_be
8667 8.3356 no-vmlinux /no-vmlinux
2263 2.1765 sbcenc sbc_encode
696 0.6694 libc-2.10.1.so memcpyBecause of the use of --separate=kernel option, we can see ~8% of cpu time attributed to no-vmlinux image, which is the time spent in the kernel mostly doing input/output activity (reading the input file from disk). Also less than 1% is spent in memcpy function which belongs to libc-2.10.1.so shared library. Without --separate=kernel option, this information would not be present in the log.
Now our focus is on sbc_calc_scalefactors_j function, which got 21548 oprofile samples collected, and they represent ~20.7% of time spent in 'sbcenc' process. Please note again, that this percentage would not be a realistic estimate without also having kernel and libc information in the picture. In the case if the CPU consumption is dominated by the library functions or by the kernel, the statistics could be severely skewed.
After performing the optimizations, we get a new profiling report:
samples % image name symbol name
26234 29.9625 sbcenc sbc_pack_frame
20057 22.9076 sbcenc sbc_analyze_4b_8s_neon
14306 16.3393 sbcenc sbc_calculate_bits
9866 11.2682 sbcenc sbc_enc_process_input_8s_be
8506 9.7149 no-vmlinux /no-vmlinux
5219 5.9608 sbcenc sbc_calc_scalefactors_j_neon
2280 2.6040 sbcenc sbc_encode
661 0.7549 libc-2.10.1.so memcpyWhich shows that now we have sbc_calc_scalefactors_j_neon function taking 5219 samples instead of 21548 samples for sbc_calc_scalefactors_j earlier. It is approximately ~4.1x speedup for this particular function. Samples are more important than percents in the log, because the absolute number of samples represents the actual time spent in the function, and the percents are relative to the whole process (as the whole program takes less time to execute after optimization, the percents may naturally drift).
For another example we can look at the sbc_pack_frame function statistics in both logs. The number of samples remained about the same: 26083 vs. 26234 (see the 3-sigma rule from the 'A bit of theory' section). But the percentage of the time relative to the whole program increased from ~25% to ~30% even though this function has not changed itself. That's a nice side affect of optimizations: after eliminating the obvious bottlenecks, the other functions are becoming more attractive optimization targets too :)
The precision of measurements can be always increased by running the test program more than one time between 'opcontrol --start' and 'opcontrol --stop' invocations, because more samples will get accumulated and the relative error will become smaller.
Still the other methods of benchmarking the code may be more suitable for very tiny performance tweaks, such as just saving maybe a few CPU cycles. Some tricks for benchmarking small sequences of instructions are described in my older Discovering instructions scheduling secrets blog post.
ARM Cortex-A8 performance monitoring unit disaster
If you tried to follow the instructions described above, but got bizarre results, then the chances are quite high that you are using some hardware with ARM Cortex-A8 processor. The problem is that ARM Cortex-A8 has a broken performance monitoring unit (this is described as erratum #628216 in ARM Cortex-A8 errata list). Earlier revisions were badly broken. The last revisions are a bit better, but still not suitable for use with oprofile.
For collecting samples, oprofile relies on the interrupts generated by the performance monitoring unit. The interrupts are supposed to happen on overflows of the 32-bit hardware performance counters. But with the older ARM Cortex-A8 revisions (for example, used in beagleboard), the PMU state may be occasionally messed up on the counter overflow. With the newer ARM Cortex-A8 revisions (for example, used in beagleboard-xm), the counter may just overflow without triggering an interrupt. The outcome is disasterious in both cases. Skipped interrupt may be difficult to notice because it takes slightly more than 4 seconds to count from 0 to 0xFFFFFFFF on a 1GHz processor. So the performance monitoring unit recovers itself, but each skipped interrupt results in approximately 4 seconds dropped out from the profiling session. Longer profiling runs have a higher chance of triggering this hardware bug eventually. And considering that it is important to collect really a lot of samples for getting good precision, Cortex-A8 performance monitoring unit using cycle counter is a really bad option.
The solution for all these troubles is simple: use the timer interrupt, Luke :) Hardware performance counters are actually more like a red herring. Timer interrupt works perfectly fine for the simple profiling tasks, so there is no point trying to use the performance monitoring unit no matter what. Admittedly, I have wasted quite a lot of time myself trying to workaround this pesky issue.
In order to override Cortex-A8 performance monitoring unit with a simple timer driver, adding "oprofile.timer=1" to the kernel command line can be used. Or using "timer=1" module parameter if oprofile is built as a module.
Also when using the simple timer driver, it makes sense to tweak it a bit if we don't want to collect samples at something like a pitiful default 128 Hz rate. The following hack can be applied to the linux kernel to solve this:
diff --git a/drivers/oprofile/timer_int.c b/drivers/oprofile/timer_int.c
index 3ef4462..56fb6c3 100644
--- a/drivers/oprofile/timer_int.c
+++ b/drivers/oprofile/timer_int.c
@@ -20,13 +20,15 @@
#include "oprof.h"
+#define OPROFILE_TIMER_TICK_NSEC 244141 /* ~4096 Hz */
+
static DEFINE_PER_CPU(struct hrtimer, oprofile_hrtimer);
static int ctr_running;
static enum hrtimer_restart oprofile_hrtimer_notify(struct hrtimer *hrtimer)
{
oprofile_add_sample(get_irq_regs(), 0);
- hrtimer_forward_now(hrtimer, ns_to_ktime(TICK_NSEC));
+ hrtimer_forward_now(hrtimer, ns_to_ktime(OPROFILE_TIMER_TICK_NSEC));
return HRTIMER_RESTART;
}
@@ -40,7 +42,7 @@ static void __oprofile_hrtimer_start(void *unused)
hrtimer_init(hrtimer, CLOCK_MONOTONIC, HRTIMER_MODE_REL);
hrtimer->function = oprofile_hrtimer_notify;
- hrtimer_start(hrtimer, ns_to_ktime(TICK_NSEC),
+ hrtimer_start(hrtimer, ns_to_ktime(OPROFILE_TIMER_TICK_NSEC),
HRTIMER_MODE_REL_PINNED);
}Additional verification for the Poisson based stddev estimate (added on 2011-08-28)
Let's take the following profiling session as an example:
Profiling through timer interrupt
samples % image name symbol name
52105 40.0715 djpeg jpeg_idct_islow
41281 31.7473 djpeg ycc_rgb_convert
15126 11.6327 djpeg decode_mcu
15001 11.5366 djpeg h2v1_fancy_upsample
2029 1.5604 djpeg decompress_onepass
1470 1.1305 libc-2.12.2.so memset
1118 0.8598 no-vmlinux /no-vmlinux
967 0.7437 libc-2.12.2.so _wordcopy_fwd_dest_aligned
333 0.2561 djpeg jpeg_fill_bit_buffer
69 0.0531 libc-2.12.2.so fwrite
69 0.0531 libc-2.12.2.so writePoisson gives us a theoretical estimate for the standard deviation as the square root of the number of samples. But just to be sure, we can verify it by running the same profiling session 10 times and calculating sample standard deviation for the number of samples attributed to each function.
| function | time spent in the function, measured in oprofile samples | mean | sample stddev | sqrt(mean) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| jpeg_idct_islow | 52105 | 52171 | 51968 | 52243 | 52389 | 52126 | 52347 | 52217 | 52078 | 52543 | 52218.7 | 169.2 | 228.5 |
| decode_mcu | 15126 | 15119 | 15315 | 15060 | 15108 | 15397 | 15227 | 15017 | 15175 | 15138 | 15168.2 | 115.8 | 123.2 |
| decompress_onepass | 2029 | 2042 | 2070 | 2012 | 2057 | 2127 | 2022 | 2074 | 2048 | 1992 | 2047.3 | 37.98 | 45.25 |
| fill_bit_buffer | 333 | 311 | 333 | 311 | 334 | 297 | 309 | 309 | 336 | 304 | 317.7 | 14.63 | 17.82 |
By comparing the last two columns in the table, we can see that the values there are reasonably close to each other. So assuming a stable test environment with no background activity from the other processes, etc., we can run just one profiling session and already have a good estimate for the precision of the measurement for each function. Still it is a good idea to repeat profiling at least one more time and check if the results are consistent between runs in order to rule out any possible interference from the external factors or the problems in the whole setup (see the 'ARM Cortex-A8 performance monitoring unit disaster' section). If the results are not consistent across runs, it makes sense identifying and eliminating the source of this noise.
Also the applicability of the Poisson based standard deviation estimate is limited to the functions which take a reasonably small percentage of time (as wikipedia article says: "The Poisson distribution can be applied to systems with a large number of possible events, each of which is rare. A classic example is the nuclear decay of atoms"). And if taking a corner case as an example, if oprofile log shows that all the samples belong only to a single function ('main'), then the precision of this measurement would be very high and only depend on the timer resolution. The number of samples would be equal to the time taken by the process multiplied by the oprofile samples collection frequency. But on a positive side, sqrt(N) still provides a reliable pessimistic estimate, with the real standard deviation being lower than that.